Network Modeling
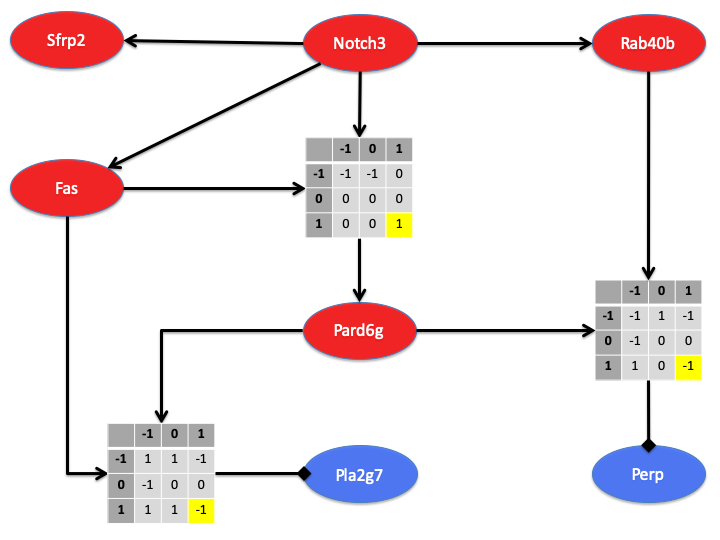
Genes do not act in isolation; rather they act together in complex networks that govern cellular function. Investigation of the interactions between genes (and gene products) is necessary to elucidate cellular mechanisms. Disease progression produces drastic changes in genetic networks critical to normal cellular function. Accurate models of the gene regulatory networks present in diseased and normal cells can further our understanding of the disease and potentially inform both prognosis and treatment.
Modeling of gene regulatory networks is inherently difficult for four reasons: (1) high order interactions are common, (2) dependencies are often non-linear, (3) there are numerous unmeasured variables, and (4) biological measurements are noisy and prone to technical artifacts. We have developed novel methodology to estimate gene regulatory networks that addresses each of these challenges. Specifically, we have developed an approach to network estimation that includes non-linear multi-gene interactions, is agnostic with respect to unmeasured variables, and explicitly models and incorporates uncertainty in each step of the analysis. Quantifying model uncertainty allows researchers to determine highly probable features and areas in which additional experimentation is needed.
The overall goal of this project is to move beyond descriptive models of gene regulatory networks and develop methods that produce testable hypotheses regarding cellular function.