Cell Morphology
Advances in high-throughput cell imaging techniques provide an opportunity to develop computational methods to jointly model cell morphology and gene expression. We developed a method to identify genes associated with context-dependent alterations in cell morphology (Nassiri and McCall, 2018). Application of this method to molecular perturbation data from the Library of Integrated Network-based Cellular Signatures (LINCS) Project identifid gene sets associated with individual morphological features.
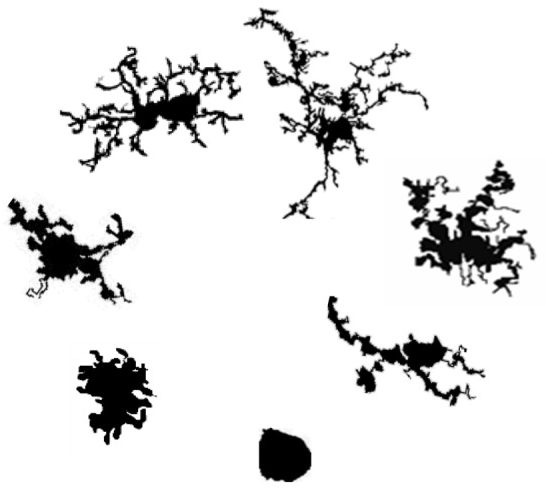
This project seeks to improve the analysis of cell morphology, specifically microglia morphology, by developing statistical methods to quantify images of cells and to use multiple measures to examine differences in broad domains of morphology between experimental conditions. The majority of current image summaries are not based on an explicit statistical model and lack a corresponding measure of uncertainty. Finally, individual quantitative summaries are typically analyzed separately, ignoring the dependence between different aspects of cell morphology and thereby reducing statistical power. We are developing statistical methods to model cell morphology that fully leverage the wealth of information present in the imaging data and provide interpretable parameter estimates and corresponding measures of uncertainty.