Cell Mixtures
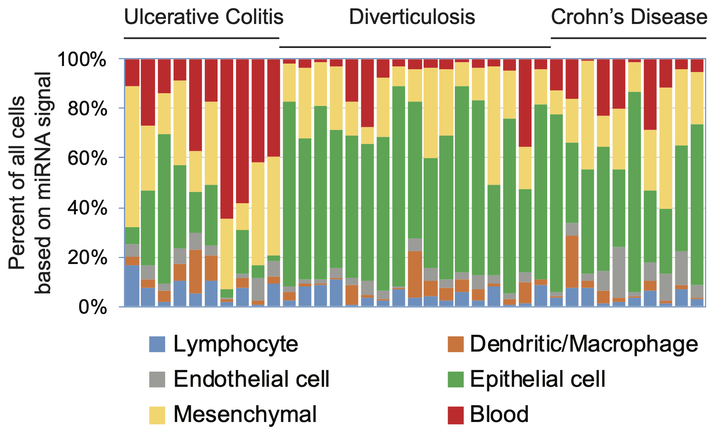
Within a tissue, component cell types have both shared and unique genomic profiles and corresponding cellular functions. We hypothesize that the changing ratios of cell types in disease are a major, underappreciated cause of tissue-level expression changes detected by standard tests for differential expression. The goal of this research is to develop computational methods to decompose a tissue-level signal into its intrinsic cell types. These methods will be used to adjust for the impact of changing cell ratios on genomic measurements within tissue samples. By adjusting for changes in cellular composition, one can isolate changes in cellular regulation and function. This work has the potential to fundamentally improve genomic analysis so that well-designed studies will identify biologically relevant changes.
Drs. Halushka and McCall jointly lead a multidisciplinary team focused on developing novel methodological and technological advances in the analysis of human tissue samples. This work has already led to several important discoveries. First, we showed that across tissues endothelial cells have both shared and unique expression patterns. Second, our investigation of miR-143/145 expression in colon tissue demonstrated importance of measuring cell type specific expression in each cell type within the system of study. Third, we uncovered extensive signal heterogeneity in nominally normal lung tissue samples from the Genotype-Tissue Expression (GTEx) project that was the result of sampling- or treatment-related changes in the cellular composition of the tissue. Most recently, we examined the effect of variation in tissue composition on estimation of gene co-expression and showed that current methods of deconvolution are only able to partial recover cell type specific co-expression.